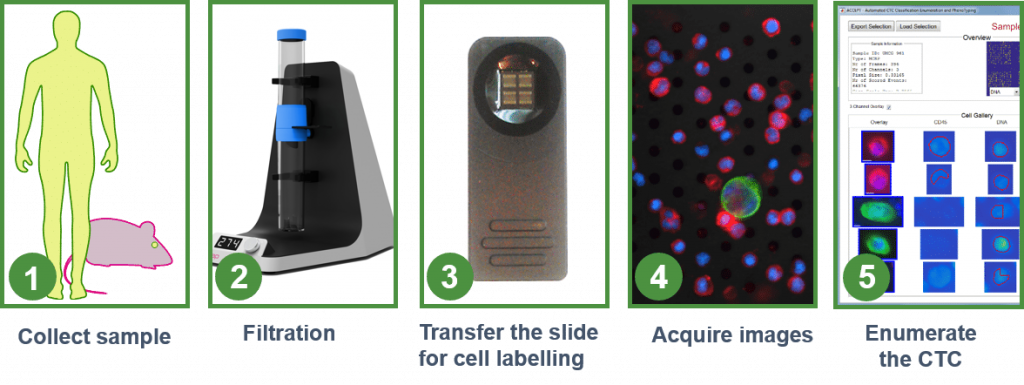
VyCAP’s CTC enumeration solution is simple to use, requires limited hands-on time and results in high recoveries. This size based filtration uses the larger size and rigidity of circulating tumor cells compared to other blood cells.
The CTC enumeration technology consists of :
- A filtration disposable, that contains a microsieve filter with defined pores (5.0 ± 0.2µm)
- An imaging system, to automatically image and enumerate CTC
VyCAP offers a complete solution for the enumeration of CTC in blood of cancer patients and xenograft models. The solution consists of hardware, software and protocols. Protocols to enumerate CTC in whole blood of xenografts and humans are present on our website.
Unique Selling Points CTC enumeration
- Short (<1hr) and simple workflow to enumerate CTC from whole blood
- High CTC recoveries >70%
- Validated protocols for Transfix (Cytomark) and CellSave (Silicon Biosystems)
- Perfect images without auto-fluorescence
- Dedicated imaging system CTC enumeration
Short and validated workflow for CTC enumeration
The workflow to collect and enumerate CTC for whole blood of human or xenograft models uses validated protocols. It takes less than 1 hr to process the sample and enumerate the circulating tumor cells. For the collection of the blood we advise to use Transfix or Cellsave blood collection tubes. All our staining and filtration reagents used in the protocols are commercially available. Contact us or download the protocols for more information.
Filtration disposable contains a filtration chip with precisely defined pores
CTCs are larger and more rigid compared to blood cells. The difference between the CTC and blood cells, like leukocytes, is however small. This requires that the pores in the filter are very precise with a minimum variation between the pores. The microsieve chip is manufactured using state of the art micro machining technologies. The microsieve filter chip contains 160.000 precise pores (pore diameter 5 ± 0.2 µm).
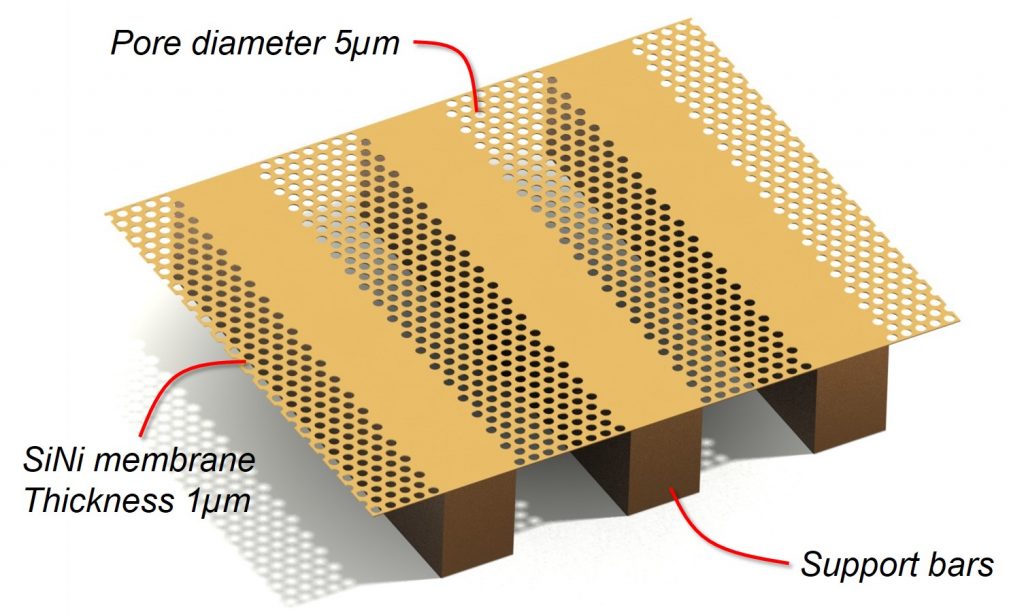
Thin microsieve membrane results in less force on cells
The silicon nitride (SiNi) filter membrane has a thickness of 1µm which is supported by a silicon structure below the membrane. Because of this very small thickness, the required pressure across the microsieve filter is much lower compared to other size based filtering systems. A low pressure results in less force and less cell damage on the captured cells.
Pump unit to filter whole blood
The microsieve filter is mounted in a plastic slide that is mounted in the filtration disposable. The disposable clicks onto the pump unit. Whole blood is transferred to the sample side of the disposable and a small negative pressure is applied by the pump unit. The used pressure is depending on the type of blood collection tube, see our protocols. It takes typically a few minutes to filter a tube of whole blood with a volume of 10 ml. Next, the slide is removed and the collected cells are immunofluorescently labelled.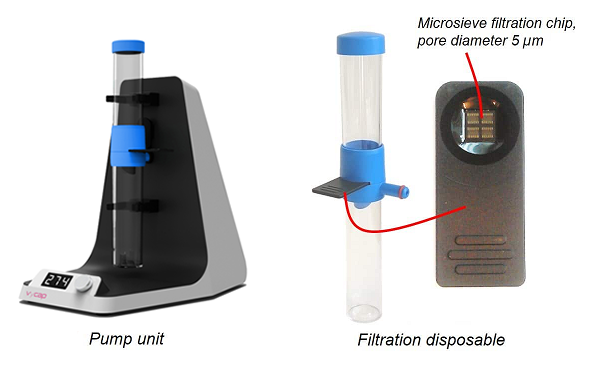
Easy cell labeling with the Cellstainer
After the filtering the slide is removed from the disposable filtration unit and placed into the cellstainer. Next, the labeling reagents are applied to the cells that are presented on the filtration membrane (Step 1). The staining reagents stay on the filter and the cells remain submerged during incubation (Step 2). To remove excess reagents, the microsieve filter is pushed onto an absorbing pad that is positioned below the microsieve filter (Step 3). This process can be repeated multiple times with different reagents. The cells will remain on the sieve and will not be washed away during the staining steps.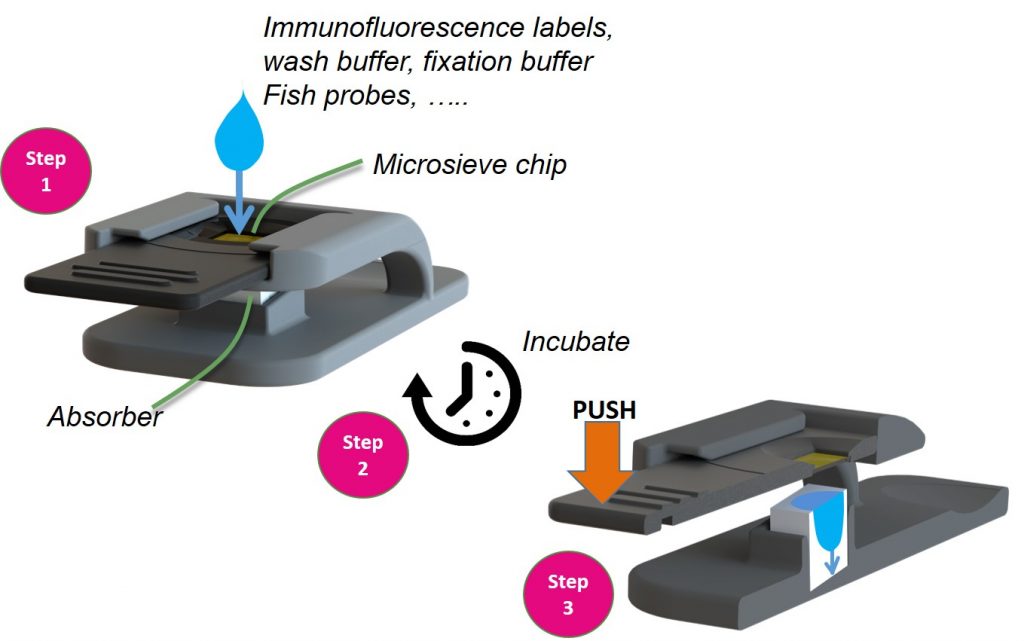
Small microsieve chip for fast imaging and image processing
Our microsieve filter chips are only 8x8mm which allows fast imaging and image processing by VyCAP software and ACCEPT. Besides the relative small surface of the microsieve filter, the membrane is atomically flat and presents no auto-fluorescence. The CTC selection software automatically selects the CTCs. After imaging, the possible CTC are presented to the user in a gallery. Standard protocols for CTC enumeration based on the DNA+, CK+, CD16- and CD45- are available and additional labels can be added (for example PDL-1 or MUC-1).
Imaging system to enumerate CTC on the microsieve
Automatically enumerate CTC with the VyCAP Imaging system. With this dedicated imaging system and software, slides with microsieves can be automatically imaged and processed. The imaging system contains high end scanning stages, a Lumencor LED excitation light source and a Hamamatsu CMOS scientific camera. The stages are automatically controlled by the image acquisition software; furthermore the system has auto focus and an automatic filter cube changer that can hold a maximum 6 different filter cubes. Up to eight slides can be loaded at once. Standard protocols for CTC counting based on the DNA+, CK+, CD16- and CD45- are available and additional labels can be added (for example PDL-1 or MUC-1). Scanning a microsieve in 4 fluorescence colors takes around 15 minutes. The same microscope is used as the base in the Puncher system and the imaging system can be upgraded to a Puncher system at any time.
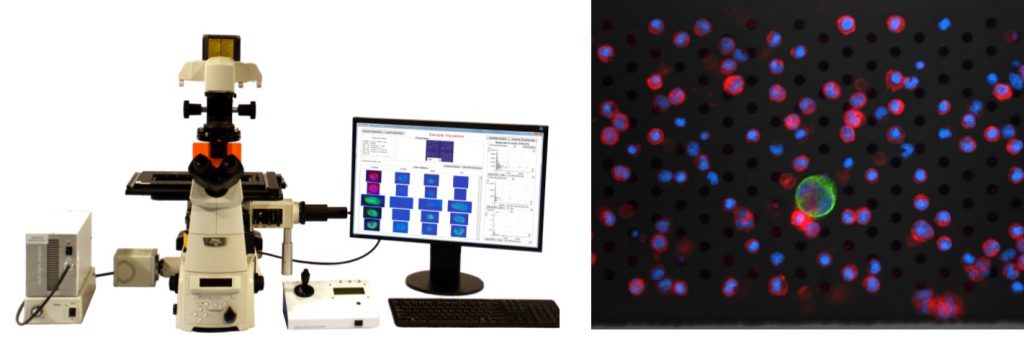
Compatible with ACCEPT for CTC detection
The images can be processed by the open source CTC image analysis program ACCEPT. This program automatically identifies CTC on different parameters using sophisticated image analysis algorithms. After processing a list of possible circulating tumor cells are presented in an image gallery. The final selection of circulating tumor cells is done by the user.
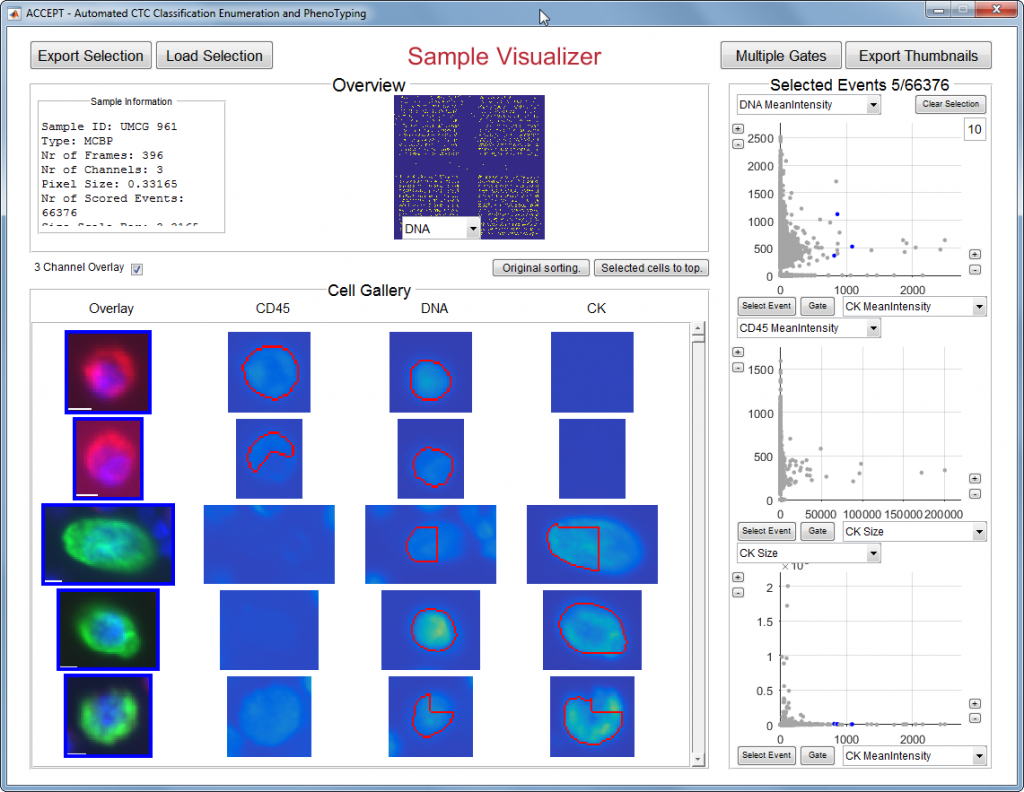
The software ACCEPT is developed by researchers in a EU-consortium called CancerID. The Public-Private-Partnership with currently 36 partners from 13 countries aims at the establishment of standard protocols for and clinical validation of blood-based biomarkers. It brings together experts from academic and clinical research, innovative Small-to-Medium sized Enterprises (SMEs), diagnostics companies and the pharmaceutical industry.