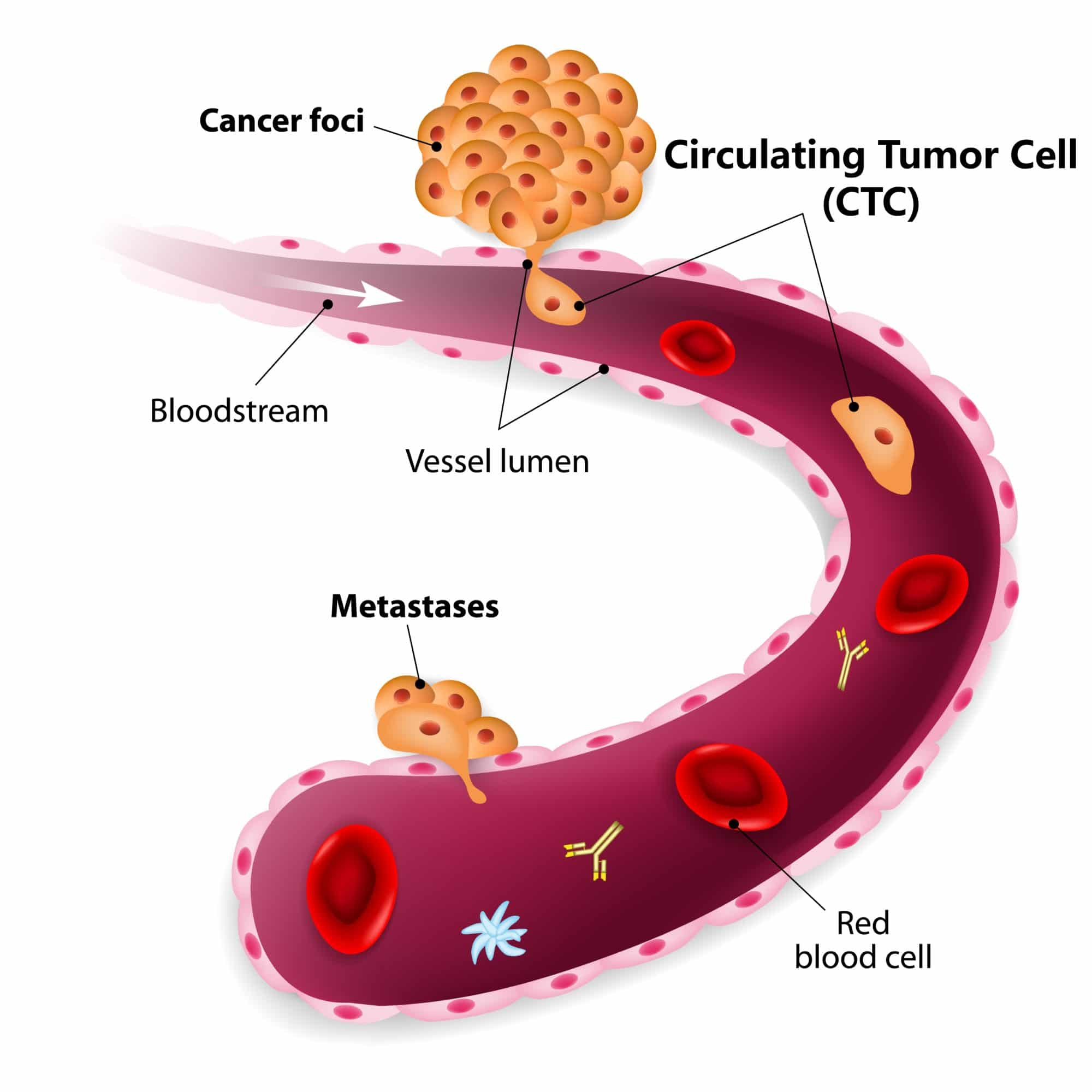
Automatically enumerate Circulating Tumor Cells (CTCs) with our complete solution. VyCAP offers a size based technology to enumerate CTC from liquid biopsies such as:
- Blood
- Urine
- Pleural effusion
- Spinal fluids
Enumeration and analysis of CTC in liquid biopsies
Enumerate Circulating Tumor Cells easily and reliably with limited hands-on time. The size based filtration uses the larger size and higher rigidity of circulating tumor cells compared to other blood cells. Epcam positive and Epcam negative cells will be captured on the microsieve. In the Cellsearch system only Epcam positive cells will be captured.
To enumerate CTC researchers need:
- A filtration disposable, that contains a microsieve filter with defined pores (5.0 ± 0.2µm)
- An imaging system, to automatically image and enumerate CTC
VyCAP offers a complete, simple, validated and automated solution to enumerate CTC in the blood of cancer patients. The system is easy and flexible. Additional labels can be added to the cells which is ideal for research.
Workflow to enumerate Circulating Tumor Cells
The workflow to enumerate CTCs in human whole blood samples is validated, automated and simple. It takes less than 1 hr to process a sample and enumerate the circulating tumor cells. Our protocols for labeling are based on commercially available reagents. If you wish to use a different immuno-fluorescence label, the protocol can easily be modified for your purpose.
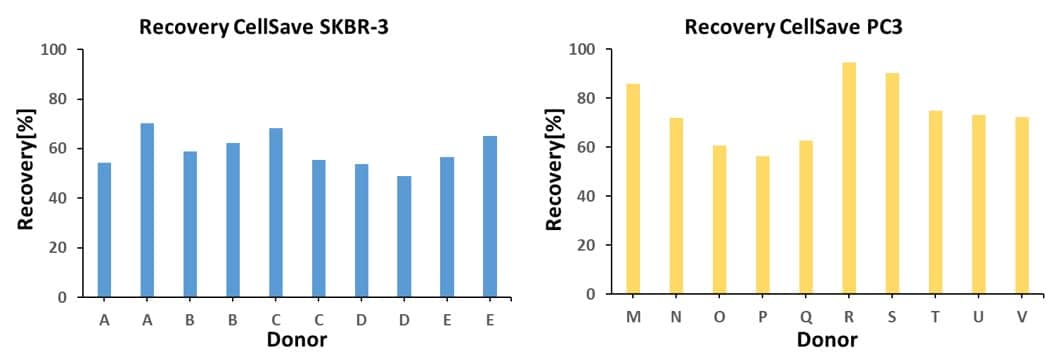
High recovery of Circulating Tumor Cells
The graphs below present the recoveries of SKBR-3 and PC-3 cells spiked in CellSave blood of healthy donors. Blood is filtered using VyCAP’s enumeration disposable and imaged and enumerated using VyCAP’s imaging system.
Enumerate CTC in blood
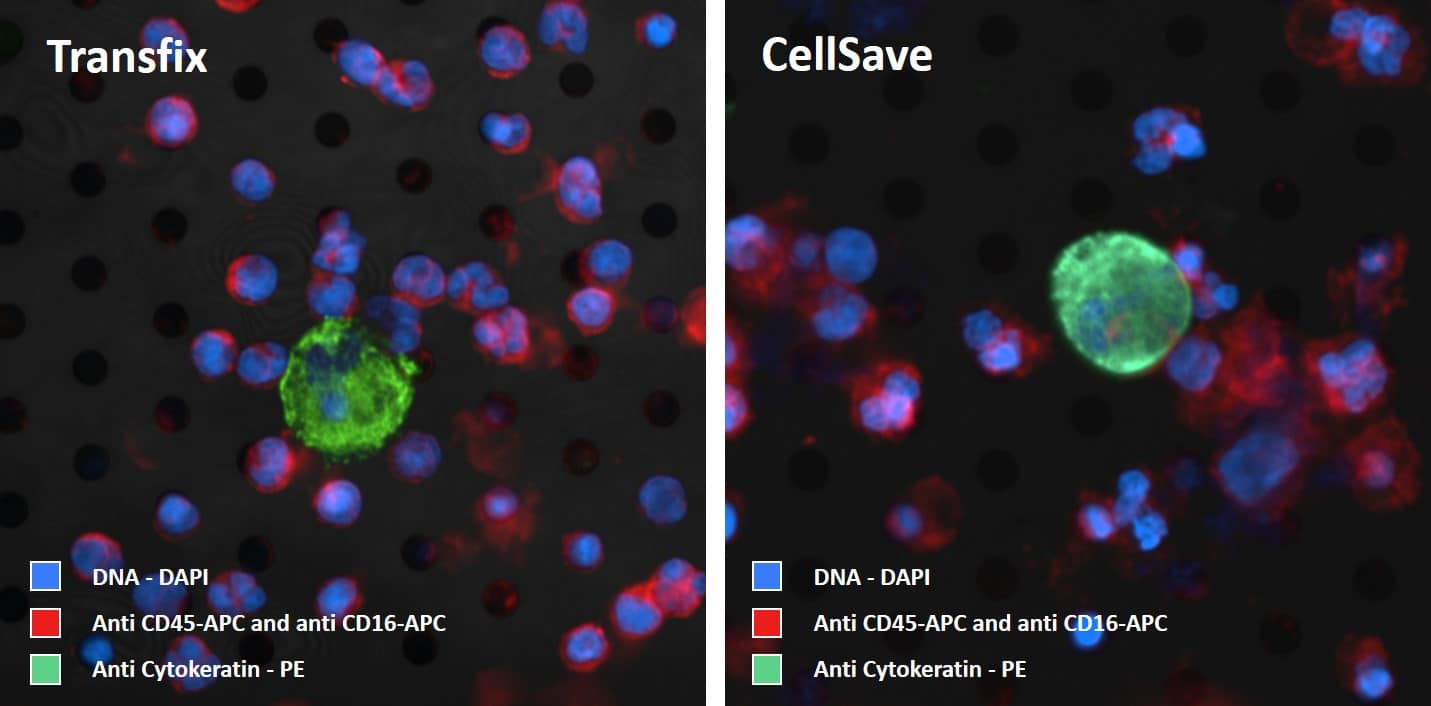
The acquired blood sample needs to be processed within 48 hrs to guarantee proper filtration. After the blood is filtered an additional 2 ml of PBS is added to rinse most of the red blood cells from the microsieve filter. The slide with microsieve filter is transferred to the cellstainer for labeling. A combination of fluorescent labels is used. Our protocol for CTC enumeration uses a combination of fluorescent labels:
- Anti Cytokeratines – Phycoerythrin (PE)
- Anti CD45 – Allophycocyanin (APC)
- Anti CD16 – Allophycocyanin (APC)
- Other labels can be added if required
After labeling, mounting medium containing DAPI is added and a coverglass is applied. The coverglass is added to obtain high quality images and prevent sample degradation. Example images of labelled cells on the microsieve are depicted below. The images are acquired with VyCAP’s imaging system.
Enumerate CTC in urine
After urine collection the sample is fixed by addition of EtOH/AcOH (3:1) in a 1:1 ratio. Subsequently the urine is filtered and cells on the microsieve filter are labelled. After the cells are labelled the slide is placed on the VyCAP imaging system. The fluorescent images are acquired. The images are automatically processed to enumerate CTC using VyCAP’s software or ACCEPT.
FISH for CTC identification in urine
The presence of squamous cells in urine, that stain as well for Cytokeratin, hamper CTC enumeration based on DNA and cytokeratin alone. Therefore FISH is used to make the final identification of the circulating tumor cells. In the below images centromeric FISH probes against chromosome 1, 7, 8 and 17 were applied for CTC identification. The presence of more than two copies clearly identifies the tumor cell. Below image presents an urine sample that is fluorescently labelled for DNA and cytokertin as well as the corresponding FISH image for CEP 1, 7, 8 and 17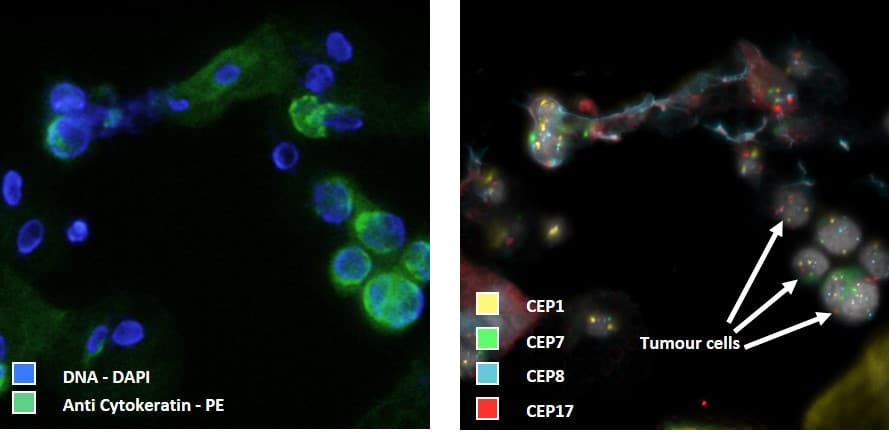
Enumerate CTC in Pleural Fluid
 Pleural fluid was obtained by a thoracentesis, an invasive procedure to remove fluid from the pleural space. The fluid was tapped from the pleural space and transferred to a 50 ml tube. To remove clots the pleural fluid was pre-filtered using a nylon mesh filter (pores of 150 µm).
Pleural fluid was obtained by a thoracentesis, an invasive procedure to remove fluid from the pleural space. The fluid was tapped from the pleural space and transferred to a 50 ml tube. To remove clots the pleural fluid was pre-filtered using a nylon mesh filter (pores of 150 µm).
Next the sample was transferred to the filtration disposable. A small negative pressure of 50 mbar was used to filter 2.5 ml of pleural fluid.
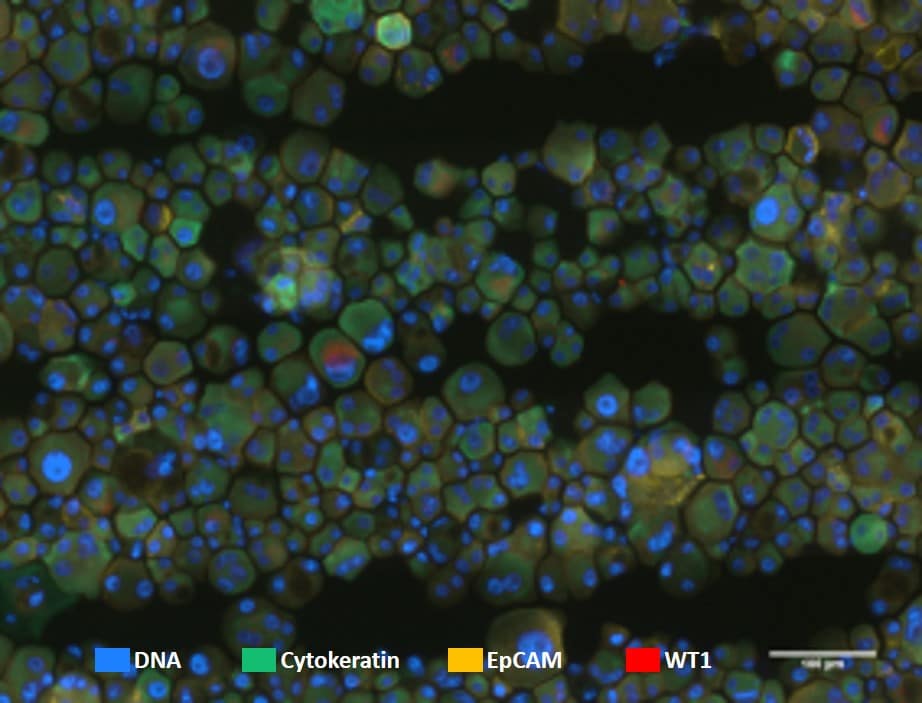
After processing the slide with the microsieve was transferred to VyCAP’s cellstainer for labeling. A panel of four fluorescence labels was applied:
- DNA
- Cytokeratine – FITC
- WT-1 – APC
- EpCAM – PE
Fluorescence images of the stained cells were acquired. Images show cells on the microsieve using a 40x objective.