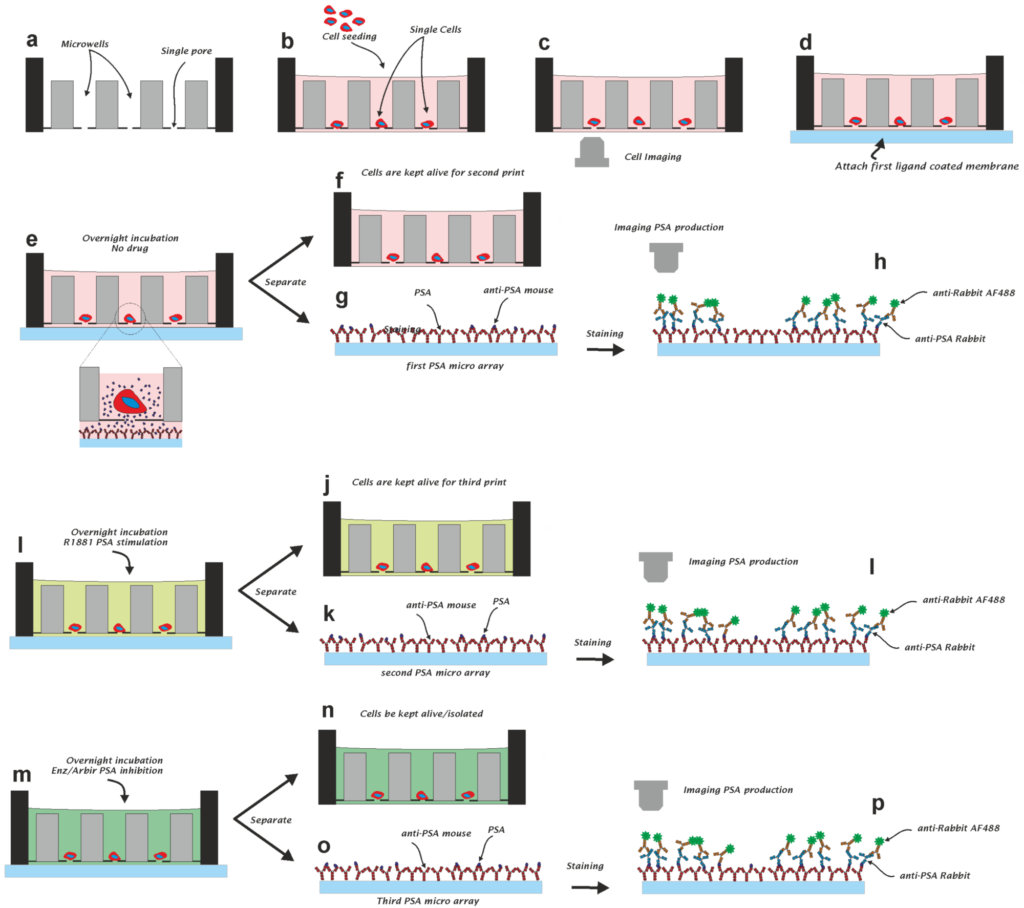
Measurement of the Drug Sensitivity of Single Prostate Cancer Cells
Fikri Abali, Narges Baghi, Lisanne Mout, Joska J. Broekmaat, Arjan G. J. Tibbe,
and Leon W. M. M. Terstappen
Abstract
Cells communicate mainly through the secretion of proteins. Impaired protein secretion can indicate the development of disease. Cancer cell heterogeneity and acquired resistance to therapy are, however, reducing the effectiveness of cancer treatments. As cancer cells change during the course of the disease, sampling of cancer cells at the time of treatment is needed in order to determine which drugs will be effective. This paper describes a method for measuring secreted prostate specific antigen (PSA) protein from thousands of prostate cancer (PCa) cells. Furthermore, we show that the PSA secretion of individual cells in microwells can be stimulated or inhibited with drugs. To this end, we believe that this method could accelerate the development of new drugs, improve our understanding of resistance to therapy, and, ultimately, improve personalized cancer therapy.
Proficiency Testing to Assess Technical Performance for CTC-Processing and Detection Methods in CANCER-ID
Rui P.L. Neves, Wim Ammerlaan, Kiki C. Andree, Sebastian Bender, Laure Cayrefourcq, Christiane Driemel, Claudia Koch, Merlin Verena Luetke-Eversloh, Marianne Oulhen, Elisabetta Rossi, Catherine Alix-Panabieres, Fay Betsou, Francoise Farace, Sabine Riethdorf, Thomas Schlange, Harriet Wikman, Rita Zamarchi, Klaus Pantel, Leon W.M.M. Terstappen, and Nikolas H. Stoecklein
Abstract
BACKGROUND: Multiple technologies are available for detection of circulating tumor cells (CTCs), but standards to evaluate their technical performance are still lacking. This limits the applicability of CTC analysis in clinic routine. Therefore, in the context of the CANCER-ID consortium, we established a platform to assess technical validity of CTC detection methods in a European multi-center setting using non-small cell lung cancer (NSCLC) as a model.
METHODS: We characterized multiple NSCLC cell lines to define cellular models distinct in their phenotype and molecular characteristics. Standardized tumor-cellbearing blood samples were prepared at a central laboratory and sent to multiple European laboratories for processing according to standard operating procedures. The data were submitted via an online tool and centrally evaluated. Five CTC-enrichment technologies were tested.
RESULTS: We could identify 2 cytokeratin expressing cell lines with distinct levels of EpCAM expression: NCI-H441 (EpCAMhigh, CKpos) and NCI-H1563 (EpCAMlow, CKpos). Both spiked tumor cell lines were detected by all technologies except for the CellSearch system that failed to enrich EpCAMlow NCI-H1563 cells. Mean recovery rates ranged between 49% and 75% for NCI-H411 and 32% and 76% for NCI-H1563 and significant differences were observed between the tested methods.
CONCLUSIONS: This multi-national proficiency testing of CTC-enrichment technologies has importance in the establishment of guidelines for clinically applicable (pre)analytical workflows and the definition of minimal performance qualification requirements prior to clinical validation of technologies. It will remain in operation beyond the funding period of CANCER-ID in the context of the European Liquid Biopsy Society (ELBS).
EMT-independent detection of circulating tumor cells in human blood samples and pre-clinical mouse models of metastasis
Jenna Kitz, David Goodale, Carl Postenka, Lori E. Lowes, Alison L. Allan
Abstract
Circulating tumor cells (CTCs) present an opportunity to detect/monitor metastasis throughout disease progression. The CellSearch® is currently the only FDA-approved technology for CTC detection in patients. The main limitation of this system is its reliance on epithelial markers for CTC isolation/enumeration, which reduces its ability to detect more aggressive mesenchymal CTCs that are generated during metastasis via epithelial-to-mesenchymal transition (EMT). This Technical Note describes and validates two EMT-independent CTC analysis protocols; one for human samples using Parsortix® and one for mouse samples using VyCap. Parsortix® identifies significantly more mesenchymal human CTCs compared to the clinical CellSearch® test, and VyCap identifies significantly more CTCs compared to our mouse CellSearch® protocol regardless of EMT status. Recovery and downstream molecular characterization of CTCs is highly feasible using both Parsortix® and VyCap. The described CTC protocols can be used by investigators to study CTC generation, EMT and metastasis in both pre-clinical models and clinical samples.
Excessive E2F Transcription in Single Cancer Cells Precludes Transient Cell-Cycle Exit after DNA Damage
Hendrika A. Segeren, Lotte M. van Rijnberk, Eva Moreno, Frank M. Riemers, Elsbeth A. van Liere, Ruixue Yuan, Richard Wubbolts, Alain de Bruin, and Bart Westendorp
Abstract
E2F transcription factors control the expression of cell-cycle genes. Cancers often demonstrate enhanced E2F target gene expression, which can be explained by increased percentages of replicating cells. However, we demonstrate in human cancer biopsy specimens that individual neoplastic cells display abnormally high levels of E2F-dependent transcription. To mimic this situation, we delete the atypical E2F repressors (E2F7/8) or overexpress the E2F3 activator in untransformed cells. Cells with elevated E2F activity during S/G2 phase fail to exit the cell cycle after DNA damage and undergo mitosis. In contrast, wild-type cells complete S phase and then exit the cell cycle by activating the APC/CCdh1 via repression of the E2F target Emi1. Many arrested wild-type cells eventually inactivate APC/CCdh1 to execute a second round of DNA replication and mitosis, thereby becoming tetraploid. Cells with elevated E2F transcription fail to exit the cell cycle after DNA damage, which potentially causes genomic instability, promotes malignant progression, and reduces drug sensitivity.
Release of Circulating Tumor Cells During Surgery for Non-Small Cell Lung Cancer: Are They What They Appear to Be?
Menno Tamminga, Sanne de Wit, Caroline Van De Wauwer, Hilda van den Bos, Joost F. Swennenhuis, Theo J. Klinkenberg, T Jeroen N Hiltermann, Kiki C Andree, Diana C.J. Spierings, Peter M Lansdorp, Anke van den Berg, Wim Timens, Leon WMM Terstappen and Harry JM Groen
Abstract
Purpose: Tumor cells from lung cancer patients are expelled from the primary tumor into the blood, but difficult to detect in the peripheral circulation. We studied the release of circulating tumor cells (CTC) during surgery to test the hypothesis that CTC counts are influenced by hemodynamic changes (caused by surgical approach) and manipulation. Experimental Design: Patients undergoing video-assisted thoracic surgery (VATS) or open surgery for (suspected) primary lung cancer were included. Blood samples were taken before surgery (T0) from the radial artery (RA), from both the RA and pulmonary vein (PV) when the PV was located (T1) and when either the pulmonary artery (T2 open) or the PV (T2 VATS) was dissected. The CTC were enumerated using the CellSearch system. Single-cell whole genome sequencing was performed on isolated CTC for aneuploidy. Results: CTC were detected in 58/138 samples (42%) of 31 patients. CTC were more often detected in the PV (70%) compared to the RA (22%, p<0.01) and in higher counts (p<0.01). After surgery, the RA but not the PV showed less often CTC (p=0.02). Type of surgery did not influence CTC release. Only 6/496 isolated CTC showed aneuploidy, despite matched primary tumor tissue being aneuploid. Euploid so-called CTC had a different morphology than aneuploid. Conclusion: CTC defined by CellSearch were identified more often and in higher numbers in the PV compared to the RA, suggesting central clearance. The majority of cells in the PV were normal epithelial cells and outnumbered CTC. Release of CTC was not influenced by surgical approach.
A microwell array platform to print and measure biomolecules produced by single cells
Fikri Abali, Joska Broekmaat, Arjan Tibbe, Richard Schasfoort, Leonie Zeune and Leon Terstappen
Abstract
Here we describe a combined method to monitor the secretion of molecules produced by single cells, followed by a method to isolate the individual cells that produced these molecules. The method is based on a self-sorting microwell chip that is connected to an activated membrane that collects the produced molecules. The produced molecules are printed by diffusion in small spots onto the membrane. The location of the printed spots can be correlated to the microwell number and the cell that produced these molecules. To demonstrate the method, we used the EpCAM antibody producing hybridoma cell line VU1D9 and a genetically engineered CHO cell-line producing Her2. VU1D9 cells produced 4.6 ± 5.6 pg (mean ± SD) of EpCAM antibody per 24 h and CHO cells 6.5 ± 8.2 pg per 24 h of Herceptin antibody.
VyCAP’s Puncher Technology for Single Cell Identification, Isolation, and Analysis
Michiel Stevens, Lisa Oomens, Joska Broekmaat, Joris Weersink, Fikri Abali, Joost Swennenhuis, Arjan Tibbe
Abstract
Here we present the Puncher technology for the isolation of single cells. This technology combines a silicon chip with microwells, fluorescence imaging, and a punching method to isolate and transfer the single cells to standard reaction tubes. The technology is compatible with commercially available downstream workflows and instrumentation.
Here we focus on the isolation of CTC but the Puncher technology can be applied to isolate single cells from liquid biopsies and more general from cell suspensions. It is especially suited for cell suspensions that contain:
- Cells of interest at a frequency of 1 per 10,000 or less
- A low total number of cells ranging from 1 to 100,000, that are present in a volume of 0.01 to 50 mL.
The frequency of appearance of CTC in blood is in the order of the 1 per 106 leukocytes. To be able to isolate the single CTC with the Puncher technology, enrichment of the CTC by a 3 logs reduction of the leukocytes is required. Here we describe the use of Rosettesep and Parsortix as examples of pre-enrichment methods that are compatible with the Puncher technology and further downstream applications.
Highly multiplexed profiling of cell surface proteins on single circulating tumor cells based on antibody and cellular barcoding
Chunying Wang, Liu Yang, Zhuo Wang, Jianjun He, Qihui Shi
Abstract
Circulating tumor cells (CTCs) are extraordinarily rare in blood samples and represent a real-time Bliquid biopsy of tumors. Although genetic and transcriptional sequencing of single CTCs has been reported, these methods fail to provide phenotypic and functional information of CTCs such as protein levels of surface proteins. Studies of single-cell proteomic assays of CTCs have been rare because of a lack of single-cell proteomic methods to handle and analyze rare cells in a high background of non-target cells with high sensitivity, throughput, and multiplexing capacity. Here, we develop a microchip-assisted single-cell proteomic method for profiling surface proteins of CTCs based on antibody and cellular DNA barcoding strategy. We combine DNAencoded antibody tags and cell indexes to profile 15 proteins in ~ 100 single rare cells simultaneously, and use high-throughput sequencing as the readout to generate surface protein profiles of CTCs according to their cell indexes and antibody-derived protein barcodes. A 6400-well microchip and the automated puncher are used to rapidly retrieve single CTCs from enriched CTC population with minimal cell loss (~ 10%). This technological platform integrates reliable isolation and proteomic analysis of single CTCs and can be extendable to ~ 100 proteins in hundreds of rare cells with single-cell precision.
Self-Seeding Microwells to Isolate and Assess the Viability of Single Circulating Tumor Cells
Kiki C. Andree, Fikri Abali, Lisa Oomens, Fiona R. Passanha, Joska J. Broekmaat, Jaco Kraan, Pauline A.J. Mendelaar, Stefan Sleijfer and Leon W.M.M. Terstappen
Abstract
The availability of viable tumor cells could significantly improve the disease management of cancer patients. Here we developed and evaluated a method using self-seeding microwells to obtain single circulating tumor cells (CTC) and assess their potential to expand. Conditions were optimized using cells from the breast cancer cell line MCF-7 and blood from healthy volunteers collected in EDTA blood collection tubes. 43% of the MCF-7 cells (nucleus+, Ethidium homodimer-1-, Calcein AM+, α-EpCAM+, α-CD45-) spiked into 7.5 mL of blood could be recovered with 67% viability and these could be further expanded. The same procedure tested in metastatic breast and prostate cancer patients resulted in a CTC recovery of only 0–5% as compared with CTC counts obtained with the CellSearch® system. Viability of the detected CTC ranged from 0–36%. Cell losses could be mainly contributed to the smaller size and greater flexibility of CTC as compared to cultured cells from cell lines and loss during leukocyte depletion prior to cell seeding. Although CTC losses can be reduced by fixation, to obtain viable CTC no fixatives can be used and pore size in the bottom of microwells will need to be reduced, filtration conditions adapted and pre-enrichment improved to reduce CTC losses.
Isolation of single cells for protein therapeutics using microwellselection and Surface Plasmon Resonance imaging
F. Abali, M. Stevens, A.G.J. Tibbe, L.W.M.M. Terstappen, P.N. van der Velde, R.B.M. Schasfoort
Abstract
Here the feasibility is demonstrated that by combining Surface Plasmon Resonance Imaging (SPRi) and self-sorting microwell technology product secretion of individual cells can be monitored. Additionally isolation of the selected cells can be performed by punching the cells from the microwells using coordinates of the positions of microwells obtained with SPRi. Cells of interest can be retrieved sterile from the microwell array for further cultivation.
Liquid biopsy for monitoring anaplastic lymphoma kinase inhibitors in non-small cell lung cancer: two cases compared
Mariangela Manicone, Maria Chiara Scaini, Maria Grazia Rodriquenz, Antonella Facchinetti, Alfredo Tartarone, Michele Aieta, Rita Zamarchi, Elisabetta Rossi
Abstract
Three to seven percent of non-small cell lung cancer (NSCLC) patients show anaplastic lymphoma kinase (ALK)-translocation and could be treated with ALK-inhibitors. However, under crizotinib, a first-generation ALK-inhibitor, patients develop drug resistance after a median of 12 months. To overcome crizotinib resistance, several next-generation ALK inhibitors have been developed. In NSCLC, liquid biopsy allowed important improvements in the detection of the epidermal growth factor receptor (EGFR) mutation.
The ability of liquid biopsy to detect oncogenic gene/protein fusions is a newly investigated field, and is not routinely applied yet. We here present two NSCLC patients, both rearranged for echinoderm microtubule associated-protein like 4-anaplastic lymphoma kinase (EML4-ALK) and treated accordingly, who differed in the clinical outcome. We analyzed the predictive value of the liquid biopsy components, namely epithelial cellular adhesion molecule (EpCAM)+ circulating tumor cells (CTCs), EpCAM low/neg CTCs, EML4-ALK rearranged CTCs, and cell-free mRNA (cfmRNA), during ALK-inhibitors treatment. This analysis showed a potential association between the liquid biopsy biomarkers, patients’ outcome and response to treatment, thus suggesting their combined use in the clinical practice, as proposed here. This approach would allow longitudinal monitoring and consequent identification of putative drug-resistance mechanisms, in the light of improving high-risk patient management.
Microfluidic device for DNA amplification of single cancer cells isolated from whole blood by self-seeding microwells
Yang Y, Rho HS, Stevens M, Tibbe AG, Gardeniers H, Terstappen LW
Abstract
Self-seeding microwell chips can sort single cells into 6400 wells based on cell size and their identity verified by immunofluorescence staining. Here, we developed a microfluidic device in which these single cells can be placed, lysed and their DNA amplified for further interrogation. Whole blood spiked with MCF7 tumor cells was passed through the microwell chips after leukocyte depletion and 37% of the MCF7 cells were identified by epithelial cell adhesion molecule (EpCAM) staining in the microwells. Identified single cells were punched into the reaction chamber of the microfluidic device and reagents for cell lysis and DNA amplification introduced sequentially by peristaltic pumping of micro-valves. On-chip lysis and amplification was performed in 8 parallel chambers yielding a 10,000 fold amplification of DNA. Accessibility of the sample through the reaction chamber allowed for easy retrieval and interrogation of target-specific genes to characterize the tumor cells.
Isolation of single cells for protein therapeutics using microwell selection and Surface Plasmon Resonance imaging
F. Abali, M. Stevens, A.G.J. Tibbe, L.W.M.M. Terstappen, P.N. van der Velde, R.B.M. Schasfoort
Abstract
Here the feasibility is demonstrated that by combining Surface Plasmon Resonance Imaging (SPRi) and self-sorting microwell technology product secretion of individual cells can be monitored. Additionally isolation of the selected cells can be performed by punching the cells from the microwells using coordinates of the positions of microwells obtained with SPRi. Cells of interest can be retrieved sterile from the microwell array for further cultivation.
Self-seeding microwell chip for the isolation and characterization of single cells
Joost F. Swennenhuis, Arjan. G. J. Tibbe, Michiel Stevens, Madhumohan R. Katika, Joost van Dalum, Hien Duy Tong, Cees J.M. van Rijn and Leon W.M.M. Ter Stappen
Abstract
A self-seeding microwell chip is introduced for the isolation and interrogation of single cells. A cell suspension is transferred to a microwell chip containing 6400 microwells, each microwell with a single 5 μm pore in the bottom. The fluid enters the microwell and drags a cell onto the pore. After a cell has landed onto the pore, it will stop the fluid flow through this microwell. The remaining fluid and cells will be diverted to the next available microwell. This results in a fast and efficient distribution of single cells in individual microwells. After identification by fluorescence microscopy, the cells of interest are isolated from the microwell by punching the bottom together with the cell. The overall single cell recovery of seeding followed by isolation of the single cell, is >70% with a specificity of 100% as confirmed by the genetic make-up of the isolated cells.
The detection of EpCAM+ and EpCAM– circulating tumor cells
Sanne de Wit, Guus van Dalum, Aufried T. M. Lenferink, Arjan G. J. Tibbe, T. Jeroen N. Hiltermann, Harry J. M. Groen, Cees J. M. van Rijn and Leon W. M. M. Terstappen
Abstract
EpCAM expressing circulating tumor cells, detected by CellSearch, are predictive of short survival in several cancers and may serve as a liquid biopsy to guide therapy. Here we investigate the presence of EpCAM+ CTC detected by CellSearch and EpCAM– CTC discarded by CellSearch, after EpCAM based enrichment. EpCAM– CTC were identified by filtration and fluorescent labelling. This approach was validated using different cell lines spiked into blood and evaluated on blood samples of 27 metastatic lung cancer patients. The majority of spiked EpCAM+ cells could be detected with CellSearch, whereas most spiked cells with EpCAMlow or EpCAM– expression were detected using filtration. Five or more CTC were detected in 15% of the patient samples, this increased to 41% when adding the CTC detected in the discarded blood. The number of patients with CTC and the number of CTC detected were doubled by the presence of EpCAM– CTC. In this pilot study, the presence of EpCAM+ CTC was associated with poor outcome, whereas the EpCAM– CTC were not. This observation will need to be confirmed in larger studies and molecular characterization needs to be conducted to elucidate differences between EpCAM– and EpCAM+ CTC.
Feasibility of a simple microsieve-based immunoassay platform
D.R. Zweitzig, Arjan G.J. Tibbe, A.T. Nguyen, C.J.M. van Rijn, M.J. Kopnitsky, K. Cichonski, Leonardus Wendelinus Mathias Marie Terstappen
Abstract:
The intrinsic properties of silicon microsieves, such as an optically flat surface, high overall porosity, and low flow resistance have led to an increasing number of biotechnology applications. In this report, the feasibility of creating a microsieve-based immunoassay platform was explored. Microsieves containing 5 μm pores were coupled with poly-acrylic acid chains, and then mounted into a plastic holder to enable rapid reagent exchanges via a wicking mechanism. The mounted microsieves were coated with infectious disease-related antigens at [2.5 and 25 μg/mL], [20 and 50 μg/mL], and [20 and 100 μg/mL] to facilitate detection of serum-derived human antibodies against Rubella (3-day measles), B. burgdorferi (Lyme disease), or T. pallidum (syphilis), respectively. The prototype microsieve-based immunoassay platform was able to distinguish positive control sera containing antibodies against Rubella, T. pallidum, and B. burgdorferi from negative control sera with similar qualitative results as FDA-approved ELISA tests. Testing of a WHO IgG syphilitic standard at 0.3, 0.15, 0.075, 0.0375, and 0.01875 IU/mL demonstrated that the T. pallidum microsieve assay is able to distinguish disease-specific IgG signal from background signal at similar, and possibly lower, levels than the corresponding ELISA. The T. pallidum microsieve assay prototype also differentiated positive clinical serum samples from negative donor samples, and the results were in good agreement with ELISA (R2 = 0.9046). These feasibility studies demonstrate the potential for utilizing microsieves, along with a reagent wicking device, as a simple diagnostic immunoassay platform
Filter Characteristics Influencing Circulating Tumor Cell Enrichment from Whole Blood
Frank A. W. Coumans , Guus van Dalum , Markus Beck, Leon W. M. M. Terstappen
Abstract
A variety of filters assays have been described to enrich circulating tumor cells (CTC) based on differences in physical characteristics of blood cells and CTC. In this study we evaluate different filter types to derive the properties of the ideal filter for CTC enrichment. Between 0.1 and 10 mL of whole blood spiked with cells from tumor cell lines were passed through silicon nitride microsieves, polymer track-etched filters and metal TEM grids with various pore sizes. The recovery and size of 9 different culture cell lines was determined and compared to the size of EpCAM+CK+CD45−DNA+ CTC from patients with metastatic breast, colorectal and prostate cancer. The 8 µm track-etched filter and the 5 µm microsieve had the best performance on MDA-231, PC3-9 and SKBR-3 cells, enriching >80% of cells from whole blood. TEM grids had poor recovery of ∼25%. Median diameter of cell lines ranged from 10.9–19.0 µm, compared to 13.1, 10.7, and 11.0 µm for breast, prostate and colorectal CTC, respectively. The 11.4 µm COLO-320 cell line had the lowest recovery of 17%. The ideal filter for CTC enrichment is constructed of a stiff, flat material, is inert to blood cells, has at least 100,000 regularly spaced 5 µm pores for 1 ml of blood with a ≤10% porosity. While cell size is an important factor in determining recovery, other factors must be involved as well. To evaluate a filtration procedure, cell lines with a median size of 11–13 µm should be used to challenge the system.
Filtration Parameters Influencing Circulating Tumor Cell Enrichment from Whole Blood
Coumans, F. A. W., van Dalum, G., Beck, M., & Terstappen, L. W. M. M.
Abstract:
Filtration can achieve circulating tumor cell (CTC) enrichment from blood. Key parameters such as flow-rate, applied pressure, and fixation, vary largely between assays and their influence is not well understood. Here, we used a filtration system, to monitor these parameters and determine their relationships. Whole blood, or its components, with and without spiked tumor cells were filtered through track-etched filters. We characterize cells passing through filter pores by their apparent viscosity; the viscosity of a fluid that would pass with the same flow. We measured a ratio of 5·104:102:1 for the apparent viscosities of 15 µm diameter MDA-231 cells, 10 µm white cells and 90 fl red cells passing through a 5 µm pore. Fixation increases the pressure needed to pass cells through 8 µm pores 25-fold and halves the recovery of spiked tumor cells. Filtration should be performed on unfixed samples at a pressure of ~10 mbar for a 1 cm2 track-etched filter with 5 µm pores. At this pressure MDA-231 cells move through the filter in 1 hour. If fixation is needed for sample preservation, a gentle fixative should be selected. The difference in apparent viscosity between CTC and blood cells is key in optimizing recovery of CTC.
Detection of Circulating Tumor Cells
Sanne de Wit, Guus van Dalum, and Leon W.M.M. Terstappen
Abstract
The increasing number of treatment options for patients with metastatic carcinomas has created an accompanying need for methods to determine if the tumor will be responsive to the intended therapy and to monitor its effectiveness. Ideally, these methods would be noninvasive and provide quantitative real-time analysis of tumor activity in a variety of carcinomas. Assessment of circulating tumor cells shed into the blood during metastasis may satisfy this need. Here we review the CellSearch technology used for the detection of circulating tumor cells and discuss potential future directions for improvements.
Unmasking circulating tumor cells
Joost Franciscus Swennenhuis
Abstract
The number of Circulating Tumor Cells (CTCs) that can be isolated from blood of cancer patients is prognostic for the course of the disease. A higher number of CTCs correlates with a worse prognosis. A change from a higher number to a lower number of CTCs indicates a benefit of the current treatment for the patient and vice versa. For this reason the number of CTCs found with the CellSearch test can be used as a tool in the management of patient treatment. Repeated molecular analysis of CTCs enables the detection of changes during the course of the disease. This analysis could unmask details on proliferation stimulating pathways in the cells, which can lead to a change to a more effective treatment. With this information, targeted drugs could effectively be used until new analysis shows resistance and discloses possible new druggable pathways. This thesis covers the whole workflow from blood draw up to the analysis of single CTC genomes. We give an overview of all work done to analyze or unmask the cells captured with CellSearch tests and a review is included of single cell isolation methods suitable for sorting CTCs. In a review of Whole Genome Amplification (WGA) methods we show the technical details of the commercially available kits and the applicability is discussed on upstream target DNA quality and downstream applications. Cancerous origin of the epithelial cells isolated with CellSearch was expected but has been conclusively proven by the development of a Fluorescence In Situ Hybridization (FISH) test. The analysis of a large number of CTC’s from a series of prostate cancer patient samples clearly shows aberrant chromosome numbers. During development of FISH using unique sequence probes a highly efficient method was developed to remove repetitive DNA from these probes. We present the procedure to remove the repetitive DNA and show the quality and efficiency of the derived FISH probes. The possibility of analyzing genomes of single CTCs after CellSearch shows to be informative but labour intensive. The presented method using Fluorescence Assisted Cell Sorting (FACS) is a multi step procedure suitable for research but needs to be optimized and simplified to be able to use in routine diagnostics. The microwell single cell isolation technique has the potential of automation, speed and still a high degree of multi marker confirmation to authenticate the CTCs. It is a promising step forward in handling and separating CTCs.
The role and definition of cancer cells in blood
Guus van Dalum
Abstract
Cancer is the leading cause of morbidity and mortality in high developed countries and will be a major cause of morbidity and mortality for the entire world in the near future. It is therefore also a disease which is and has been subject to extensive research. Cancer is slowly moving away from being lethal to becoming a disease that is treatable. Ideally all cancer types should be treatable as a chronic disease. With the increased understanding of cancer it has become clear that cancer is a very heterogeneous disease in many ways. A large number of risk factors including numerous genetic
predispositions and environmental factors have been identified. Also the cancer cells within a patient can be very heterogeneous. Because not all cells are similar, it is possible that some cells respond differently to the same treatment. This is one of the reasons that a tumor can develop drug resistance. This raises the question, which of these cell types cause the spread of cancer through the body and form distant metastasis. Not all steps in the metastatic cascade from primary tumor to the formation of a distant metastasis are fully understood. But to spread, the cells have to leave the primary tumor and enter the blood circulation. Cancer cells that spread through the lymphatic system will also end up in the blood circulation. Although technically challenging, these cancer cells in the blood stream can be isolated and are called circulating tumor cells. They can provide real time information of the metastatic process and knowledge of the composition of these tumor cells should be ideal to select the most
appropriate therapy. The presence of tumor cells in blood is known to be prognostic for poor survival in patients with disseminated disease and can be used to evaluate the response of patients to treatment. Heterogeneity of tumor cells and changes in time giving rise to resistance to therapy requires access to tumor cells throughout the course of the disease. Identification, differentiation and characterisation of cancer cells in blood is the subject
of this thesis.
Expansion of Cancer Cells in Self-Sorting Microwells
Fikri Abali
Abstract:
Metastasis is the major cause of death in patients diagnosed with invasive cancer. Blood contains two types of cancer-derived materials that are susceptible to detailed molecular analysis: intact circulating tumor cells (CTC) and (ctDNA). CTCs from peripheral blood hold important information that may represent a “liquid biopsy” and may be used for personalized treatment and real-time monitoring of cancer patients. However, the isolation and characterization of these CTCs is extremely difficult because of their rarity and heterogeneity. Although technically challenging, the growth of CTCs from patients could overcome these difficulties. In order to better understand cellular processes in metastasis and CTC heterogeneity, a controlled way for the isolation and sorting of single CTCs and their clone formation is greatly needed.
This report is composed of two parts: part 1 of this thesis describes a self sorting microfluidic chip with 6400 microwells in an array format, applied for cell culture. Obtained results showed that cells can be seeded into each microwell of the chip automatically by filtration using a small pressure, followed by week-long culture and detection of cell growth. Growth of LNCaP cell was shown to be low when compared to MCF-7 cells. In addition the growth of 3D tumor organoids in the microwells was shown to be feasible. Organoids derived from primary cells were established in microfluidic wells and maintained in culture for weeks.
Cell growth efficiency in microwells was shown to be dependent on cell-type. Primary CRC cells displayed the highest growth efficiencies. The addition of BME showed to induce cell growth in microwells. However, the growth from single cells was still very low. In contrast increasing the cell number per microwell resulted in an increase in growth efficiency for MCF-7 cells and primary CRC cells.
Subsequently, we showed that microfluidic chip enables the retrieval of cells for continued culture or analysis. Single cells or cell grown in the microwells from a single clone could be recovered from the microwells by punching. Recovered cells proliferated after punching and gave rise to larger cell colonies.